Restriction Enzyme Ecori Cuts Between
Restriction Enzyme Ecori Cuts Between. A restriction enzyme is an enzyme isolated from bacteria that cuts dna molecules at specific sequences. Restriction enzymes cut the dna into the restriction enzyme used above is called ecori.

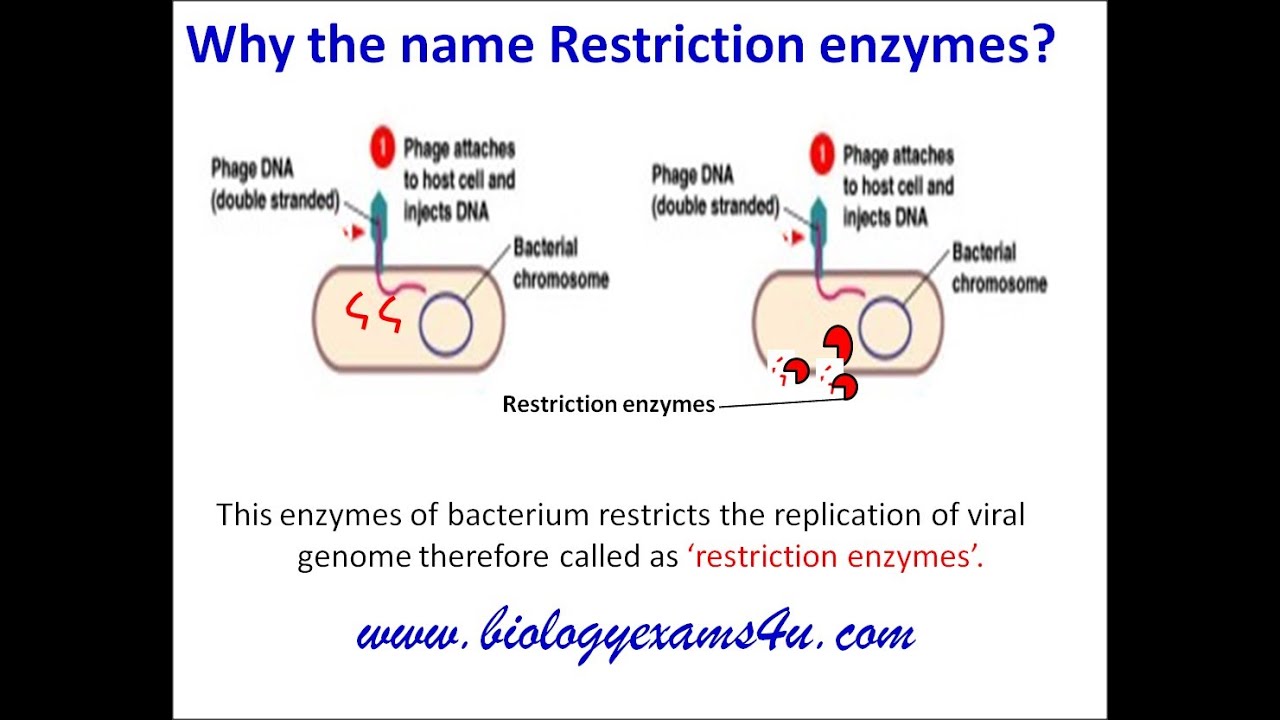
 PPT Restriction Enzymes PowerPoint Presentation, free from www.slideserve.com
PPT Restriction Enzymes PowerPoint Presentation, free from www.slideserve.comThis sequence is also a restriction site for the restriction enzyme called ecori. The key difference between ecori and hindiii restriction enzymes is that ecori is a type ii restriction enzyme that is isolated from e. ↑ where does the restriction enzyme ecori cut?

For example ecori cuts at a 6bp site, the frequency of cutting is 4^6, so it cuts every 4096bp on average, as it's sequnce will occur at random every 4096bp. Restriction enzyme or restriction endonuclease is an enzyme that cleaves dna into fragments at specific recognition.
Similarly, another restriction enzyme was named hindiii as it was isolated from haemophilus influenzae. Restriction enzymes are also known as restriction endonucleases.
Of enzymes called restriction endonucleases, or restriction enzymes. • with the aid of restriction enzymes such as ecori,
Of course, roving endonucleases can be dangerous, so bacteria protect their own dna by modifying it with methyl groups. What does r stand for in ecori?
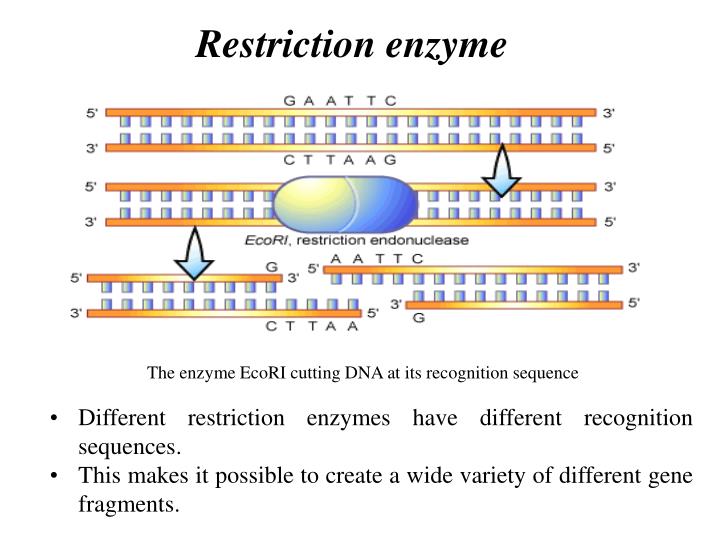
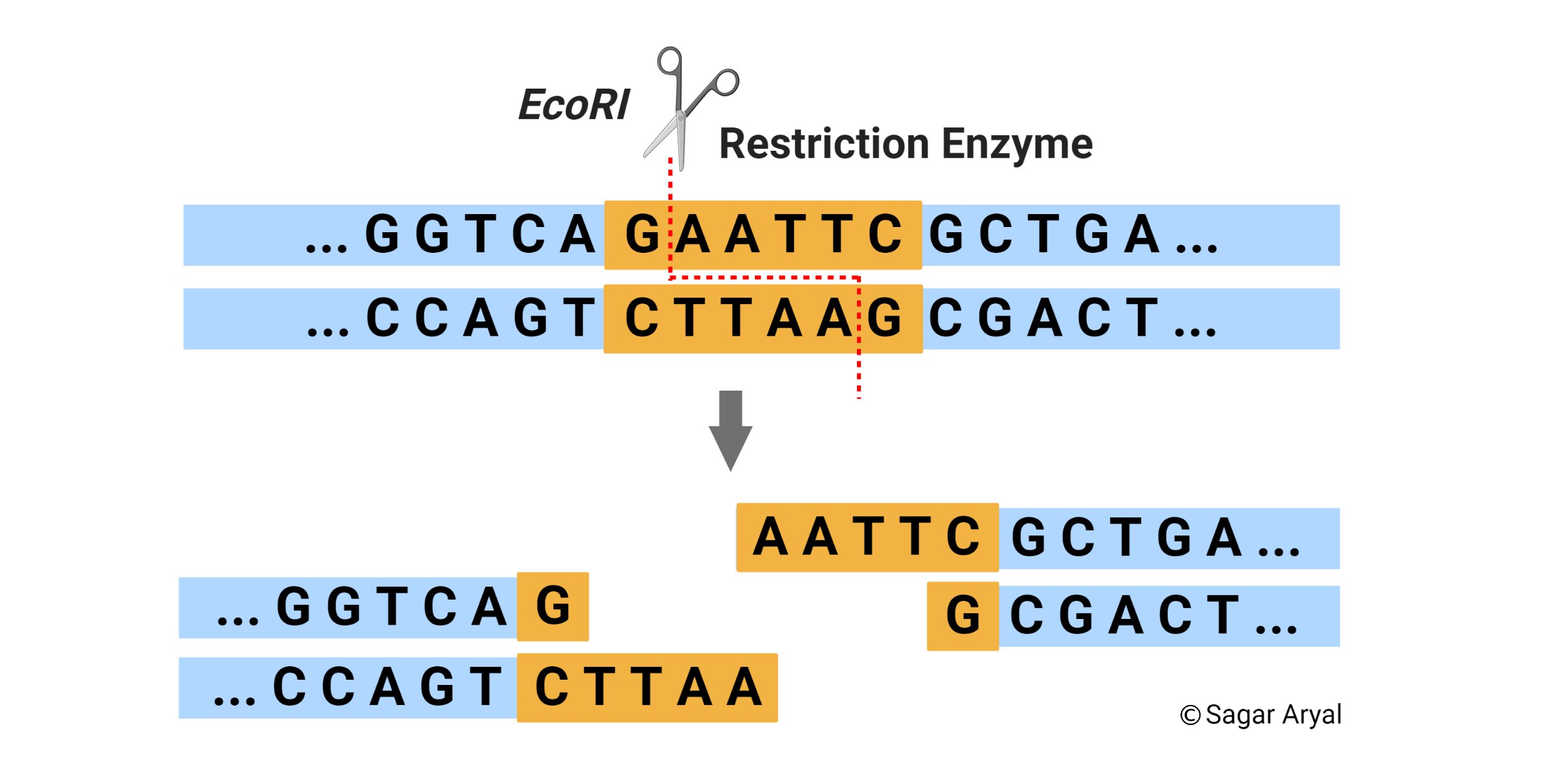
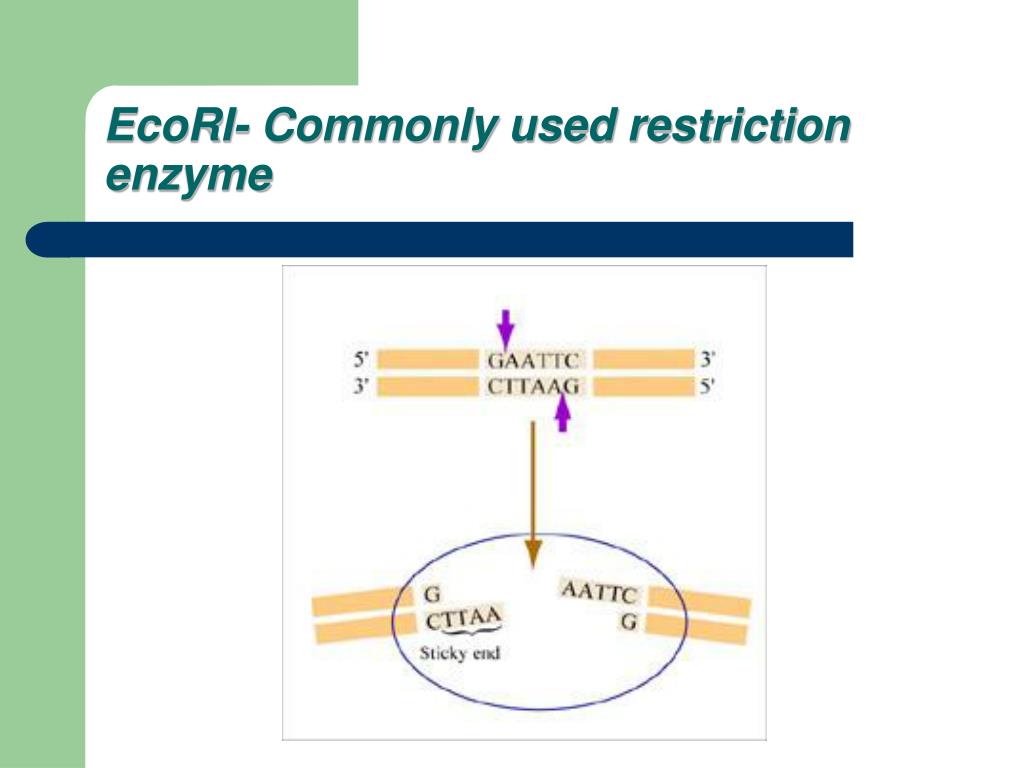
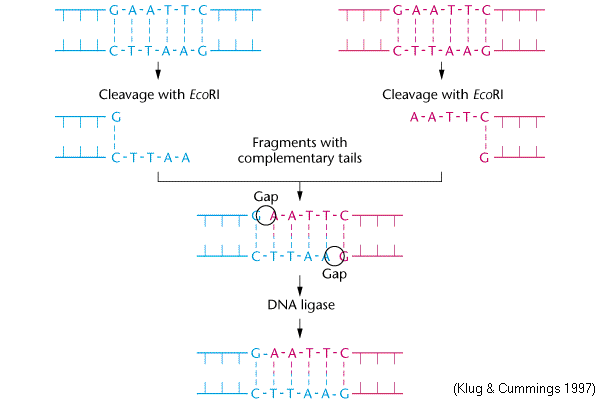
The restriction enzyme ecori cuts between g and a in the palindromic sequence gaattc. Of more value, however, are the restriction enzymes that cut between the same two bases away from the point of symmetry on two strands, thus, producing a staggering.

The majority of restriction enzymes cut 6 base pair palindromes as seen with the examples above, The chemical that cuts the dna is called a restriction enzyme.
The key difference between ecori and hindiii restriction enzymes is that ecori is a type ii restriction enzyme that is isolated from e. Of course, roving endonucleases can be dangerous, so bacteria protect their own dna by modifying it with methyl groups.
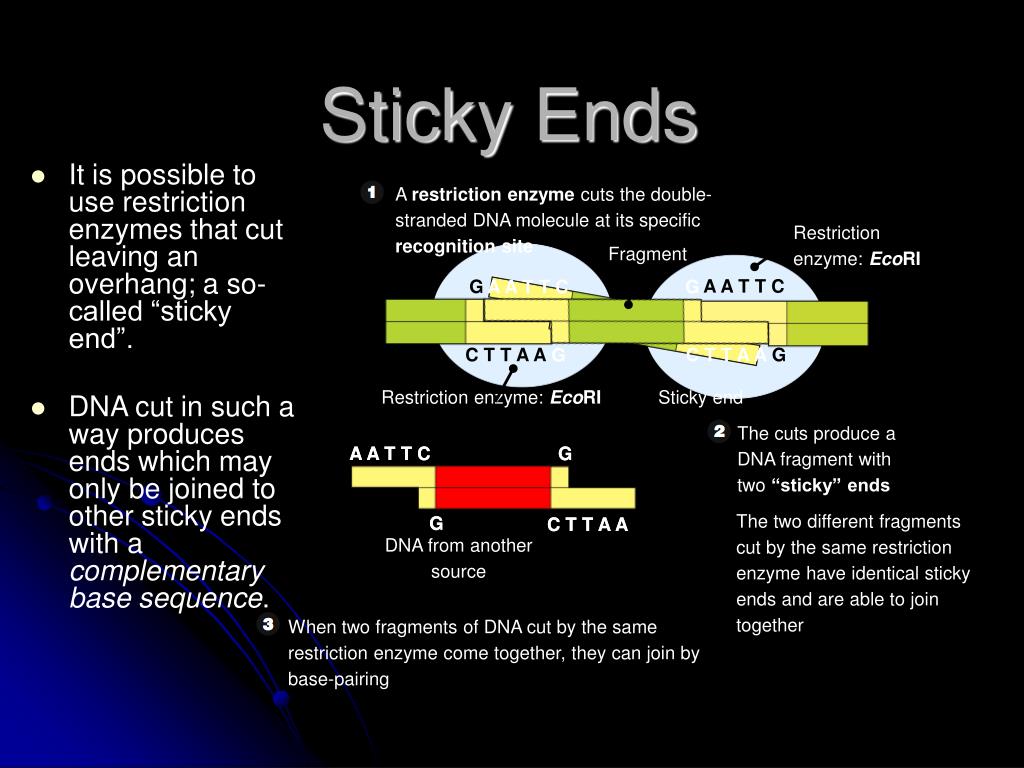
A restriction enzyme is an enzyme isolated from bacteria that cuts dna molecules at specific sequences. The restriction enzyme ecori cuts between g and a in the palindromic sequence gaattc.
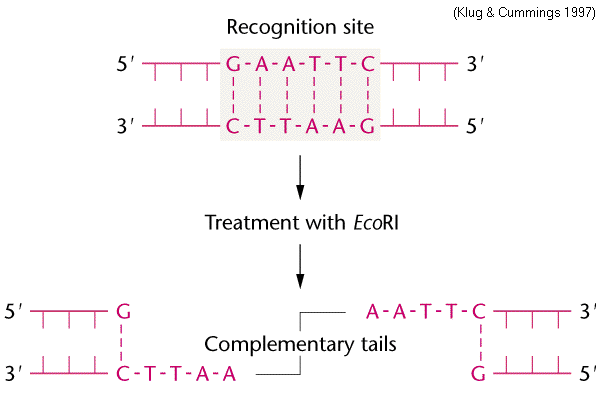
The value of restriction enzymes is that they make cuts in the dna molecule around this point of symmetry. This sequence is also a restriction site for the restriction enzyme called ecori.
The enzyme then cuts the backbones of both strands, allowing the dna to separate into two pieces. Upon restriction enzymes to cut dna at specific locations to both remove genes of interest and open chromosomes or plasmids for gene insertion.
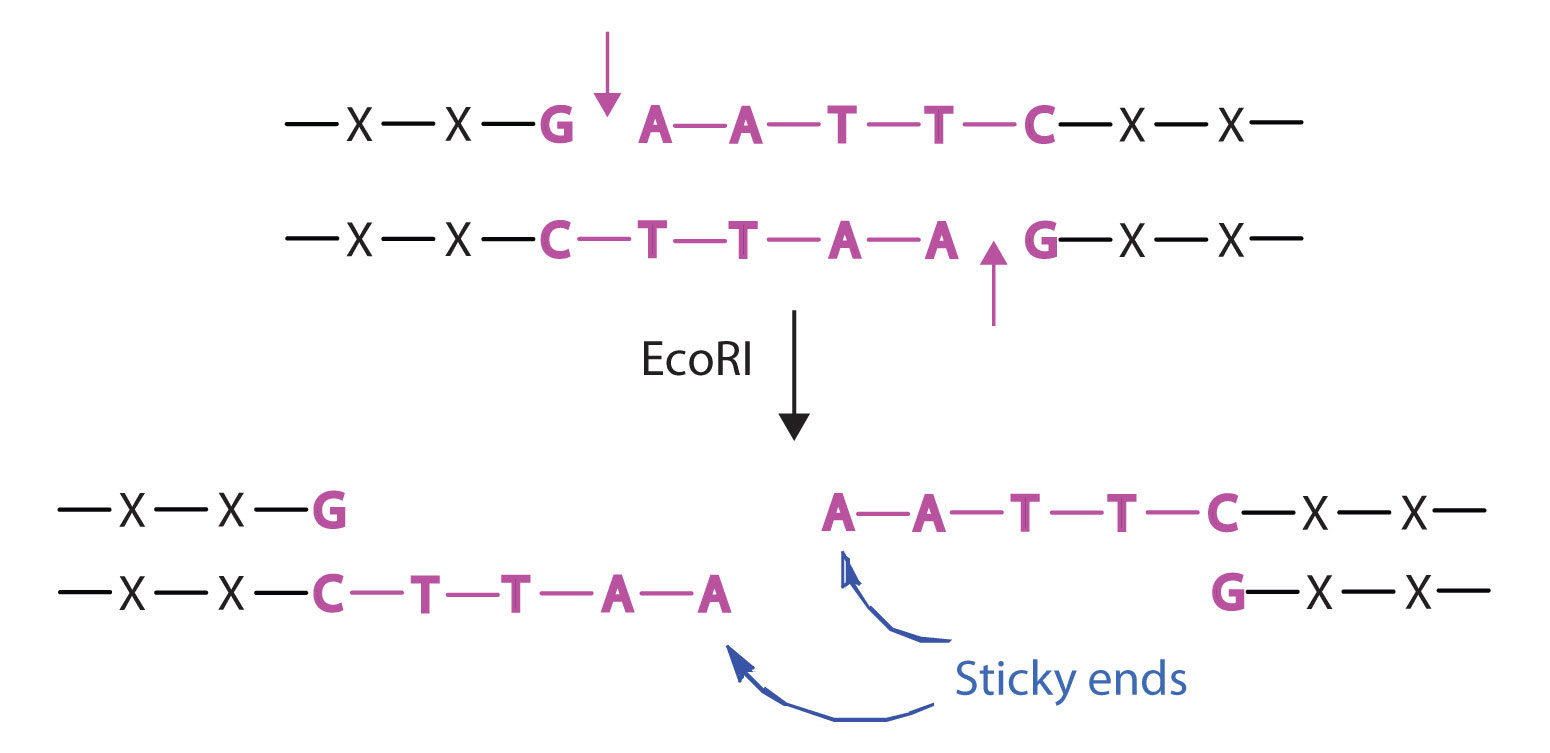
Well you can usually calculate it yourself, take the length of the genome, and divide by how often an enzyme cuts on average. Describe what a restriction enzyme does (recognize and cut at its restriction site);3.

The enzyme then cuts the backbones of both strands, allowing the dna to separate into two pieces. Each type of restriction enzyme seeks out a single dna sequence and precisely cuts it in one place.

Well you can usually calculate it yourself, take the length of the genome, and divide by how often an enzyme cuts on average. Use a restriction map to predict how many fragments will be produced in a given restrictiondigest.introductionrestriction enzymes genetic engineering is possible because of special enzymes that cut dna.

Of enzymes called restriction endonucleases, or restriction enzymes. Ecori makes one cut between the g and a in each of the dna strands (see below).
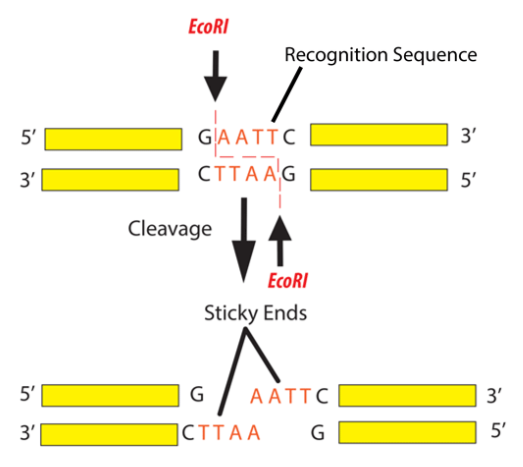
For example, the common restriction enzyme ecori recognizes the palindromic sequence gaattc and cuts between the g and the a on both the top and bottom strands. This sequence is also a restriction site for the restriction enzyme called ecori.
The enzyme then cuts the backbones of both strands, allowing the dna to separate into two pieces. While there are hundreds of different restriction enzymes, they all work in essentially the same way.
For example, the restriction enzyme ecori recognizes the palindromic sequence gaattc and cuts between the g and the a on both the top and bottom strands, leaving an overhang known as a sticky end on each end of aatt. Ecori cuts dna everywhere the base pattern copynght 1993 by trustees of boston university is found.
The name ecori comes from the bacterium in which it was discovered, escherichia coli ry 13 (ecor), and i, because it was the first restriction enzyme found in this organism. Look at the dna sequence below.
A restriction enzyme is an enzyme isolated from bacteria that cuts dna molecules at specific sequences. Ecori is a restriction enzyme or restriction endonuclease.
.jpg)
The restriction enzymes recognize short and specific nucleotide sequences in the dna known as the recognition sequences. Ecori cuts dna everywhere the base pattern copynght 1993 by trustees of boston university is found.
The Value Of Restriction Enzymes Is That They Make Cuts In The Dna Molecule Around This Point Of Symmetry.The resulting pieces of dna are typically called fragments. what makes these enzymes particularly useful is that the cut they make The enzyme then cuts the backbones of both strands, allowing the dna to separate into two pieces. Well you can usually calculate it yourself, take the length of the genome, and divide by how often an enzyme cuts on average.
While There Are Hundreds Of Different Restriction Enzymes, They All Work In Essentially The Same Way.It cuts the dna double helix at a specific site. There are 4 base pairs to the left of the cut.they are gc,ta,at,gc there are 10 base pairs to the right o. Escherichia coli strain ry13 (ecor), and i because it was the first restriction enzyme found in this organism.
For Example, The Restriction Enzyme Ecori Recognizes The Palindromic Sequence Gaattc And Cuts Between The G And The A On Both The Top And Bottom Strands, Leaving An Overhang Known As A Sticky End On Each End Of Aatt.Some restriction enzymes, for example, smai cut dna at a restriction site in a manner which leaves no overhang, called a blunt end. When the restriction enzyme recognizes a dna sequence, it hydrolyzes the bond between adjacent nucleotide and cuts through the dna molecule. For example ecori cuts at a 6bp site, the frequency of cutting is 4^6, so it cuts every 4096bp on average, as it's sequnce will occur at random every 4096bp.
Ecor1 Cuts Dna At The Recognition Sequence Between The G And A Nucleotides (See Legend For Fig.This restriction enzyme was first isolated from e.coli. • with the aid of restriction enzymes such as ecori, Because no two individuals have identical dna, no two individuals will have the same length fragments.
Restriction Enzymes Are A Class Of Enzymes That Cut Dna Into Fragments Based Upon Recognizing A Specific Sequence Of Nucleotides.Ecori is a restriction enzyme or restriction endonuclease. Look at the dna sequence below. Of course, roving endonucleases can be dangerous, so bacteria protect their own dna by modifying it with methyl groups.
Belum ada Komentar untuk "Restriction Enzyme Ecori Cuts Between"
Posting Komentar